Research
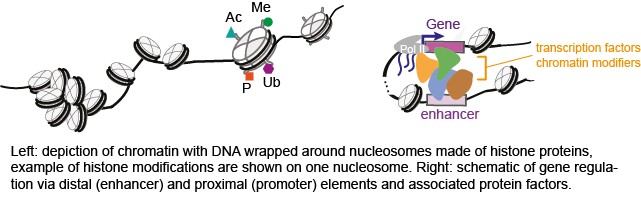
Chromatin represents the physiological form of our genome, which consists of genomic DNA closely associated with histone proteins. Post-translational modifications of the histones modulate stability and dynamics of the chromatin, thus affecting the accessibility of genomic regions to transcription factors and gene expression machinery. The precise positioning of chromatin modifications controls the activity of regulatory regions and expression of genes during cell differentiation, making epigenetic landscape one of the crucial factors in defining cell types and their developmental stages.
Mutations in chromatin-modifying proteins have been associated with the pathogenesis of both cardiovascular and neurodevelopmental disorders, pointing towards a broad role of epigenetic regulation in human pathologies. For example, up to 10% of children with congenital heart disease are also diagnosed with neurodevelopmental disorders (such as intellectual disability and autism) and carry an excess of de novo mutations related to transcriptional regulation and chromatin modifications (1). Although cardiovascular and nervous systems are highly interconnected and co-regulated, it is unclear why mutations in chromatin factors affect both systems (2). The central question of our research is how chromatin modification network affects cell differentiation and contributes to neurodevelopmental and cardiovascular disorders.
The malfunctioning of nearly all H3K4 methyltransferases (such as MLL1 and MLL2) and demethylases (LSD1) are linked to neurodevelopmental disorders (3), highlighting the significance of the H3K4-associated chromatin factors for brain function. Recently, we discovered that mutations in H3K4 demethylase LSD1, causing a novel neurodevelopmental disorder, lead to increased activity of histone deacetylases and aberrant repression of proliferation genes during cell differentiation (4). Given the high number of potential target genes of this and other chromatin factors, mis-regulation of any of them might contribute to the disease phenotype on a genome-wide scale, increasing the complexity of the perturbed cellular pathways and highlighting the need to use systemic multi-omic approaches both in bulk and in single cell to investigate these complex disease phenotypes.
Based on our recent findings and the preliminary data, we hypothesise that the mutations in chromatin modifying proteins alter the normal chromatin dynamics during cell differentiation, leading to impaired gene expression and transcription factor networks that contribute to pathogenesis in cardiovascular and neurodevelopmental diseases.
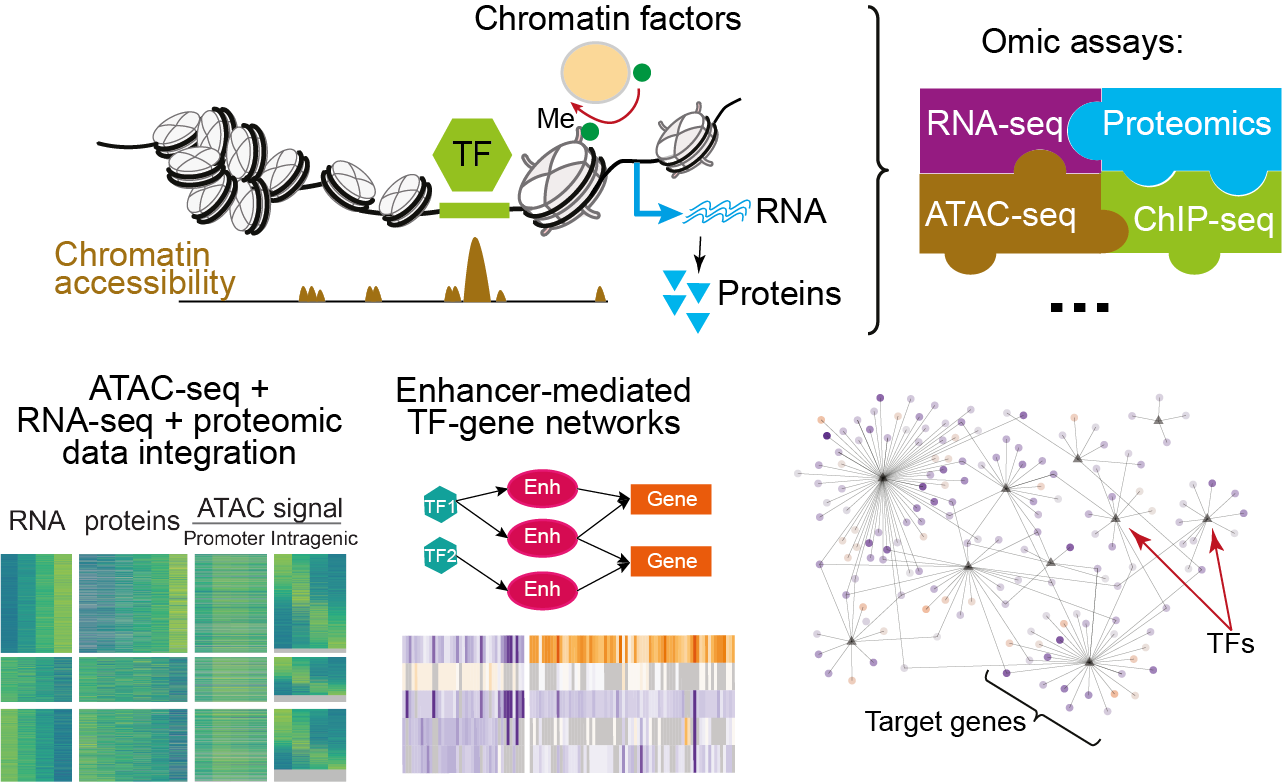
To systematically investigate which molecular pathways are affected by mutations in H3K4 modifying enzymes, we use multi-omic approaches to analyse changes in chromatin accessibility, histone modifications, activity of transcription factors, and expression of genes on RNA and protein levels using an in vitro system of iPSCs differentiation into neurons, cardiomyocytes and 3D organoid cultures. By combining the multi-omics approaches (ATAC-seq, ChIP-seq, RNA-seq and proteomics) with computational inference of TF networks (gene regulatory network analysis), we will obtain a global and systemic perspective on mis-regulated pathways caused by disease mutations. Comparing mutations in several chromatin factors causing distinct neurodevelopmental diseases with similar phenotypes, we will identify molecular pathways that are commonly perturbed in cardiovascular and neuronal development.